paired end sequencing wikipedia
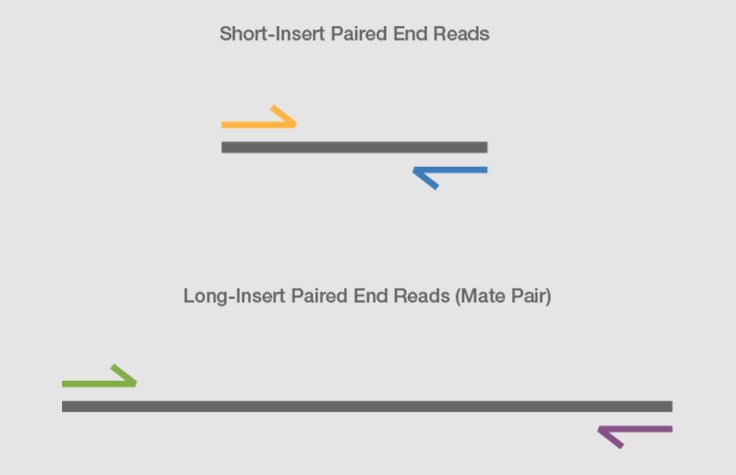
Human_only reads Aligned perfectly to human reference. Mate pair sequencing involves generating long-insert paired-end DNA libraries useful for a number of sequencing applications including.
The paired-end module of the GAII system allows both ends of a DNA fragment to be sequenced.

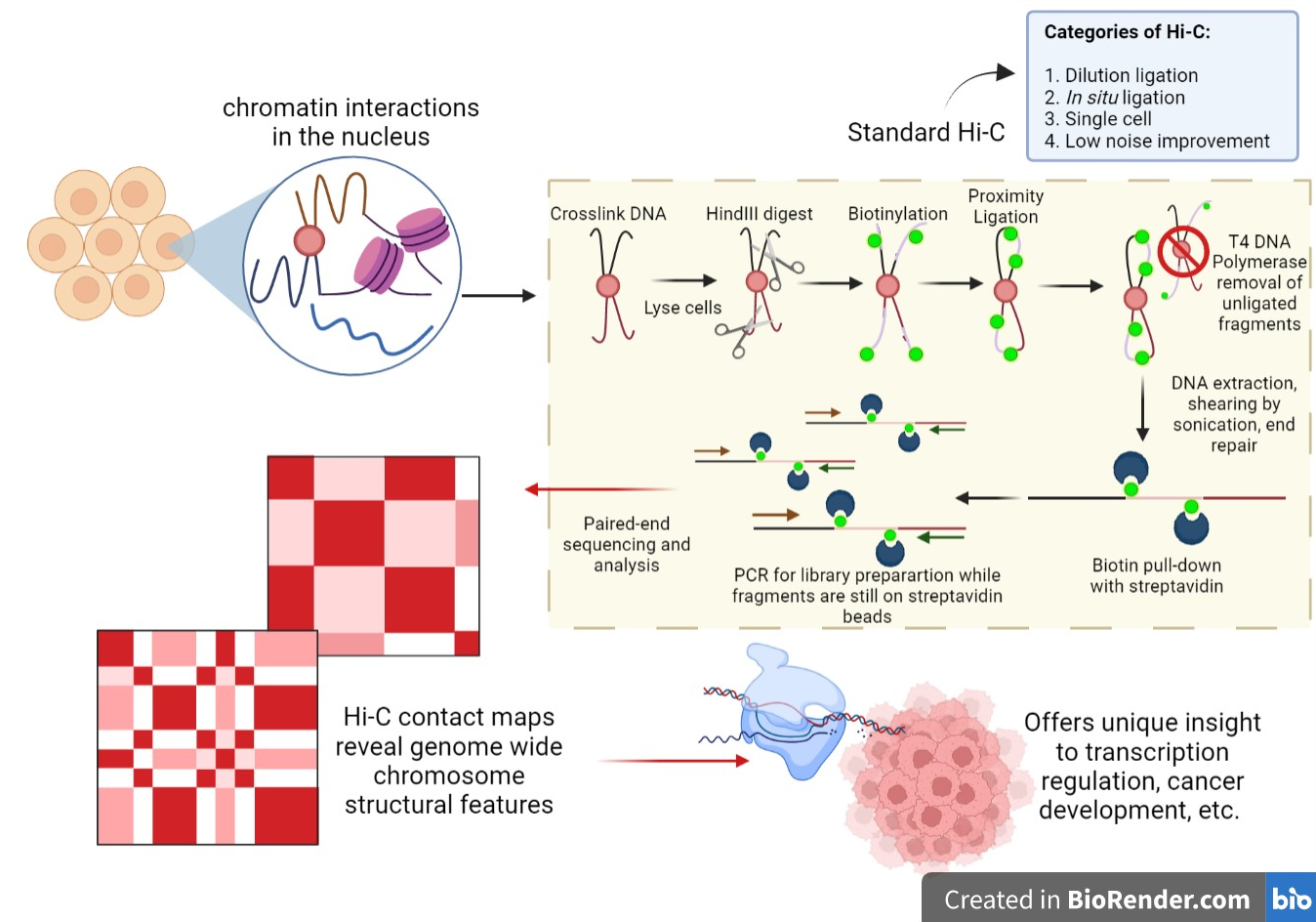
. Briefly the target genomic DNA is isolated and partially digested with restrictio. Any platform that can allow for the ligated fragments to be sequenced across the NheI junction Roche 454 or by paired-end or mate-paired reads Illumina GA and HiSeq platforms would be. Now lets get started.
Paired-end RNA sequencing RNA-Seq enables discovery applications such as detecting gene fusions in cancer and characterizing novel splice isoforms. Another consideration is whether to generate a strand-specific library that retains the orientation of the original RNA transcript which may be critical to identify antisense or non-coding RNA. As described in this wikipedia page for Illumina reads there are two possible notation for singlepaired end reads.
The figure shows the. For paired-end RNA-Seq use the. Single read runs are faster cheaper and are typically sufficient for profiling or counting studies such as RNA-Seq or ChIP-Seq.
End-sequence profiling ESP sometimes Paired-end mapping PEM is a method based on sequence-tagged connectors developed to facilitate de novo genome sequencing to identify high-resolution copy number and structural aberrations such as inversions and translocations. The inner mate distance. It is named by analogy with the rapidly expanding quasi-random shot grouping of a shotgun.
For the first test I took some sequence from the human genome hg19 and created two 100 bp reads from this region. It is important to know if the sequencing experiment was single-end or paired-end as the alignment software will. In short-read sequencing intact genomic DNA is sheared into several million short DNA fragments called reads.
Mate pair sequencing is used for various applications applications including. The program has 10 stats outputs and four BAM file outputs. A páros végű címkék PET néha páros végű diTagok vagy egyszerűen ditagok a DNS-fragmens 5 és 3 végén lévő rövid szekvenciák amelyek eléggé egyediek ahhoz hogy.
WES FAStq file is a paired end reads Stats output. The preparation of mate pair libraries is designed to allow classical paired-end sequencing of both ends of a fragment with an original size of several kilobases. These reads must first be aligned to a reference genome or transcriptome.
Massive parallel sequencing or massively parallel sequencing is any of several high-throughput approaches to DNA sequencing using the concept of massively parallel processing. Learn about the difference between Paired-End and Single-Run sequencing and why the former creates more precise alignments than the latter especiall. Paired end runs give additional positioning information in the.
Individual reads can be paired together to create paired-end. Typical experimental design advice for expression analyses using RNA-seq generally assumes that single-end reads provide robust gene-level expression estimates in a cost. Combining data generated from mate pair.
Combining data from mate pair sequencing with those from short-insert paired-end reads provides increased. Sequencing technologies vary in the length of reads produced. Paired end sequencing wikipedia Tuesday June 7 2022 Edit.
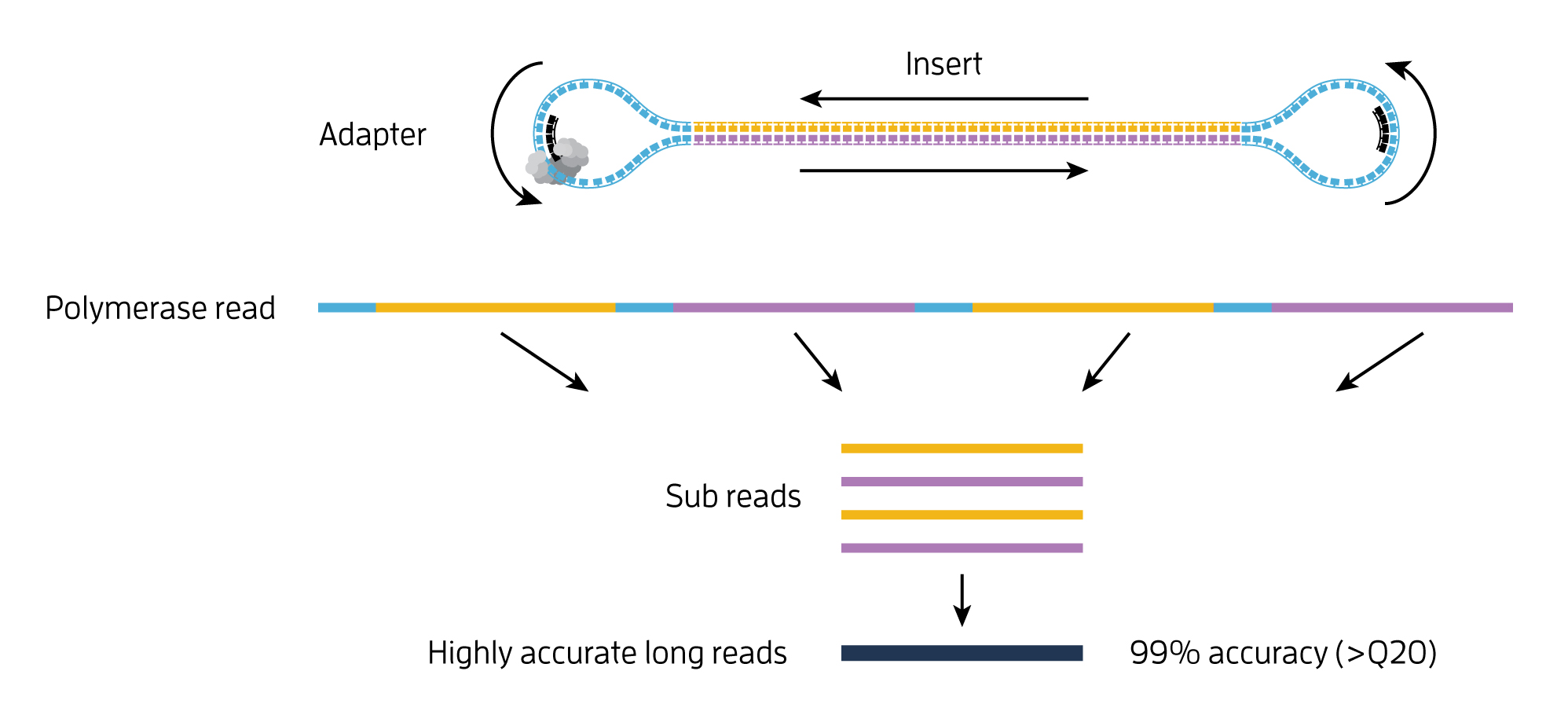
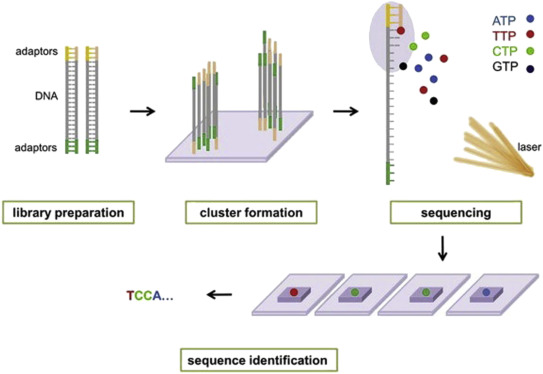
The color of the fluorescence at each interrogated position is recorded through a high-resolution camera. This gives more specificity to alignment algorithms especially in highly repetitive regions. The nanoballs are then adsorbed onto a sequencing flow cell.
In genetics shotgun sequencing is a method used for sequencing random DNA strands. First of all I checked if Sequence Id contains paired end notation. Read length is a factor which can affect the.

Maps And Rivers Middle East In 2022 Map Middle East River
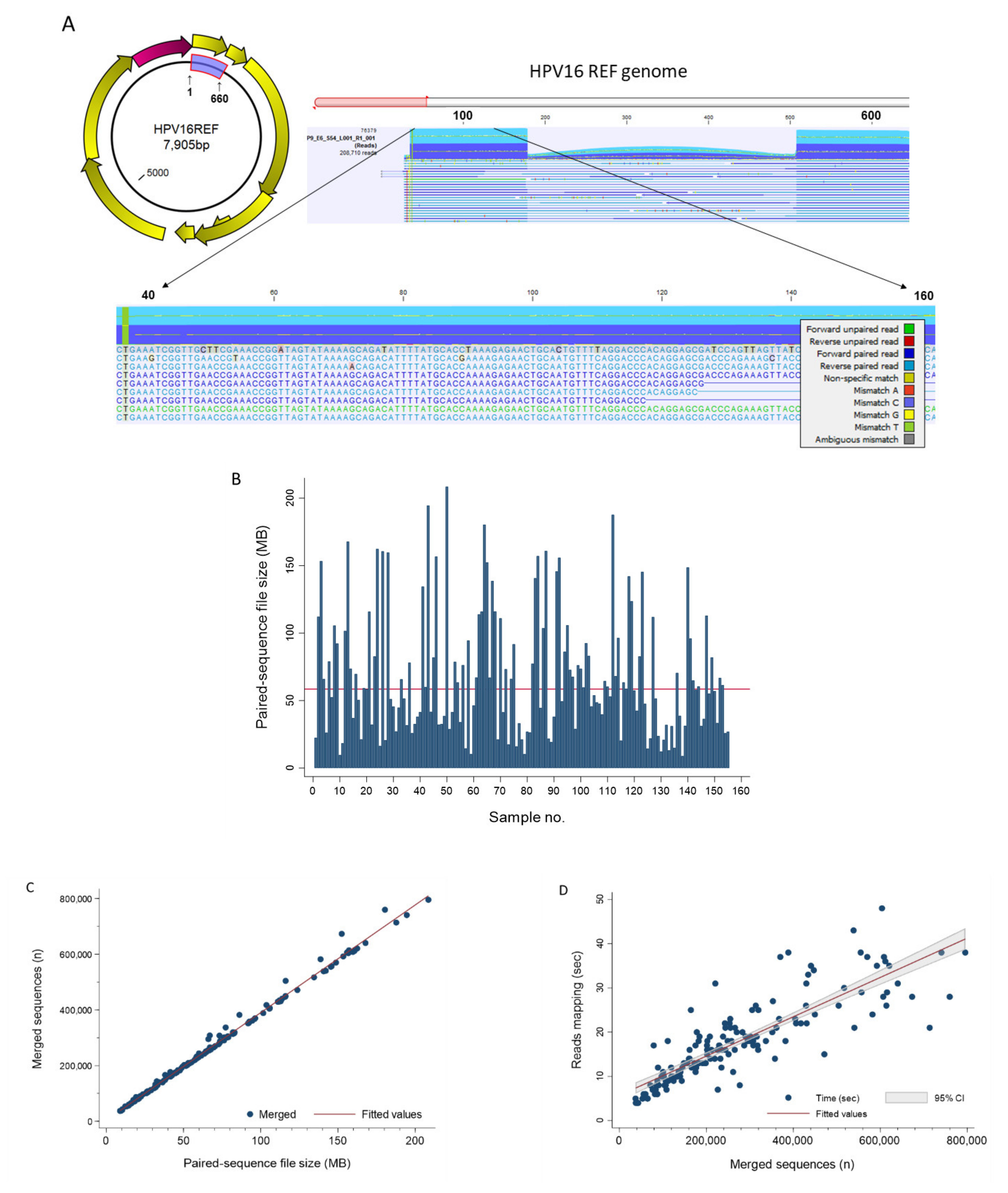
Pathogens Free Full Text Hpv Deepseq An Ultra Fast Method Of Ngs Data Analysis And Visualization Using Automated Workflows And A Customized Papillomavirus Database In Clc Genomics Workbench Html
Difference Between Whole Genome Sequencing And Shotgun Sequencing Difference Between

Restriction Site Associated Dna Sequencing An Overview Sciencedirect Topics

Bacterial Chromosome Replication Biology Lessons Chromosome Biology

What Are Paired End Reads The Sequencing Center

What Is Mate Pair Sequencing For

What Is Mate Pair Sequencing For

Bacterial Chromosome Replication Biology Lessons Chromosome Biology

What Are Paired End Reads The Sequencing Center

Paired End Shotgun Sequencing Okinawa Institute Of Science And Technology Graduate University Oist

Sequencing Introduction To Sequencing By Synthesis Sbs Youtube